Oncoscience
Abstract | PDF | Full Text | Supplementary Materials | How to Cite
https://doi.org/10.18632/oncoscience.121
TR4 nuclear receptor enhances prostate cancer initiation via altering the stem cell population and EMT signals in the PPARG-deleted prostate cells
Shin-Jen Lin1, Dong-Rong Yang1,2, Nancy Wang1, Ming Jiang3, Hiroshi Miyamoto1, Gonghui Li1,4 and Chawnshang Chang1,5
1 George Whipple Lab for Cancer Research, Departments of Pathology, Urology, Radiation Oncology, and The Wilmot Cancer Center, University of Rochester Medical Center, Rochester, NY, USA
2 Department of Urology, the Second Affiliated Hospital of Suzhou University, Suzhou, China
3 Department of Urologic Surgery, Vanderbilt-Ingram Comprehensive Cancer Center, Vanderbilt University Medical Center, Nashville, Tennessee, USA
4 Department of Urology, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China
5 Sex Hormone Research Center, China Medical University/Hospital, Taichung, Taiwan
Correspondence to: Chawnshang Chang, email: [email protected]
Correspondence to: Gonghui Li, email: [email protected]
Keywords: Prostate cancer, TR4, PPARG
Received: August 05, 2014
Accepted: February 06, 2015
Published: February 09, 2015
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
ABSTRACT
A recent report indicated that the TR4 nuclear receptor might suppress the prostate cancer (PCa) initiation via modulating the DNA damage/repair system. Knocking-out peroxisome proliferator-activated receptor gamma (PPARG), a nuclear receptor that shares similar ligands/activators with TR4, promoted PCa initiation. Here we found 9% of PCa patients have one allele of PPARG deletion. Results from in vitro cell lines and in vivo mouse model indicated that during PCa initiation TR4 roles might switch from suppressor to enhancer in prostate cells when PPARG was deleted or suppressed (by antagonist GW9662). Mechanism dissection found targeting TR4 in the absence of PPARG might alter the stem cell population and epithelial-mesenchymal transition (EMT) signals. Together, these results suggest that whether TR4 can enhance or suppress PCa initiation may depend on the availability of PPARG and future potential therapy via targeting PPARG to battle PPARG-related diseases may need to consider the potential side effects of TR4 switched roles during the PCa initiation.
INTRODUCTION
Testicular nuclear receptor 4 (TR4) belongs to the nuclear receptor superfamily and was first cloned in 1994 [1]. TR4 physiological functions are involved in metabolism, cancer development, fertility, bone diseases, cardiovascular diseases, etc [2-11]. Peroxisome proliferator-activated receptor gamma (PPARG), another nuclear receptor, also has multiple functions mostly in metabolism, cancer development, cardiovascular diseases, bone diseases, etc [12-19].
TR4 and PPARG share many similarities yet also have distinct functions in some selective diseases. First, both TR4 and PPARG genes were located in the chromosome 3p at regions 3p24 and 3p25, respectively [20]. Second, they share the same ligands/activators, i.e. polyunsaturated fatty acids and thiazolidinediones (TZDs), which transactivate their downstream target genes [11, 21-23]. Third, they bind to the similar Hormone-Response-Elements (HREs) sequences, i.e. two consecutive AGGTCA sequences with spacing of 0-5 nucleotides (direct repeat 0-5) [23]. However, more and more evidences indicate that these 2 nuclear receptors can also function oppositely in some selective diseases. First, they exert differential effects on insulin sensitivity with PPARG increasing insulin sensitivity [24-26] vs TR4 decreasing insulin sensitivity [4]. Second, PPARG suppresses atherosclerosis [17] while TR4 enhances atherosclerosis [11]. Third, PPARG increases osteoporosis [19] while TR4 decreases osteoporosis [10]. These contrasting results suggest that TR4 and PPARG may act as competitors for their upstream ligands and/or their downstream target gene modulation.
Lin et al recently reported that TR4 played a protective role in PCa initiation via modulation of the DNA damage/repair pathway [27]. They showed TR4 could suppress PCa development in 3 different mouse models and 2 different cell lines. Interestingly, Jiang et al also reported similar results showing knocking-out PPARG resulted in enhanced prostatic intraepithelial neoplasia (PIN) in the mouse model [15]. Here we report that TR4 can enhance or suppress PCa initiation depending on the availability of PPARG.
RESULTS
Selective PCa patients have one allele PPARG deletion
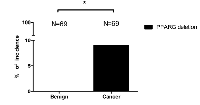
We examined PPARG deletion in PCa tissue microarray by FISH and found 9% (6 out of 69) of the PCa samples have one allele PPARG deletion (Fig. 1). In contrast, the control tissue microarray from the same patients’ normal/benign compartment has zero PPARG deletions (Fig. 1). Together, results from human clinical sample surveys suggested that PPARG deletion might be linked to the PCa development.
TR4 increases normal prostate epithelial PPARG-deleted cell growth and transformation
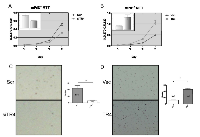
An early report suggested that PPARG had high homology with TR4 and shared similar ligands/activators [23], we were interested to see the potential differential effects of TR4 on PCa development in the normal prostate cells with or without PPARG deletion. We first applied the normal prostate epithelial mPrE-/- cell line cloned from the PPARG knockout mouse to study the TR4 effects on the PCa development. Using TR4-siRNA or TR4-cDNA to manipulate the TR4 expression, followed by the carcinogen NMU treatment to induce cell transformation [27, 28], we found knocking-down TR4 suppressed mPrE-/- cell growth using MTT assay (Fig. 2A). In contrast, addition of TR4 led to enhance mPrE-/- cell growth (Fig. 2B). Importantly, we also found knocking-down TR4 suppressed mPrE-/- cell transformation in the presence of carcinogen NMU (Fig. 2C), and addition of TR4 led to enhance mPrE-/- cell transformation using anchorage independent colony formation assay (Fig. 2D).
Interestingly, instead of knockingout PPARG, we applied the PPARG specific antagonist GW9662 [29] to suppress PPARG function, and results revealed that knocking-down TR4 led to suppress PPARG intact cell line mPrE+/+ cells treated with GW9662 (Fig. S1A). Similar results were also obtained when we replaced mPrE+/+ cells with normal human prostate epithelial RWPE1 cells (Fig. S1B).
Together, results from Fig. 2 and S1 indicated that in the prostate cells with deleted or reduced PPARG, knocking-down TR4 could suppress prostate cell growth and transformation.
TR4 enhances the PPARG-deleted prostate tumor development in the in vivo mouse model
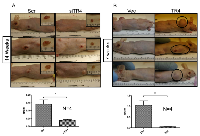
To confirm those in vitro studies showing TR4 enhanced prostate epithelial mPrE-/- cell growth and transformation in in vivo mouse model, we prepared xenografts of mPrE-/- cells with manipulated TR4 expression in nude mice. Results from Fig. 3A indicated that knocking-down TR4 led to reduced tumor sizes while over-expression of TR4 led to increased tumor sizes. Mice with TR4 knocked-down cells were sacrificed at 14 weeks after xenografts and mice with TR4 over-expressed cells were sacrificed at 7 weeks, suggesting the tumor growth was much faster in the mPrE-/- TR4 over-expressed group (Fig. 3B).
Results from Fig. 3 confirmed the above in vitro studies and concluded that TR4 might play a positive role in the PCa initiation when PPARG is deleted in prostate cells.
TR4 enhances prostate trans-differentiation in PPARG-deleted prostate cells
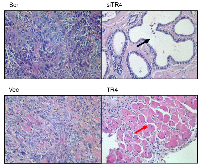
Interestingly, H&E staining of the xenografted PCa tissues prepared for Fig. 3 also found that tumors retained the adenocarcinoma structure when TR4 was knocked-down (Fig. 4). However, we found that tumors might develop into muscular structure when TR4 was over-expressed (Fig. 4). These contrasting results suggest that PCa may undergo the trans-differentiation process from the epithelial type to the mesenchymal type of cancer after TR4 was altered in the PPARG-deleted prostate cells.
TR4 increases stem cell population in PPARG-deleted prostate cells
There are several ways to trans-differentiate the epithelial cells into the mesenchymal cells [30-34]. The first way is to de-differentiate epithelial cells back into stem cells, and the resulting stem cells could then differentiate into many types of cells depending on the environment [30-32].
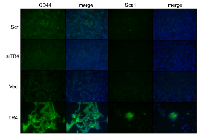
We first tested the stem cell marker CD44 and found knocking-down TR4 decreased CD44 expression while over-expressing TR4 significantly enhanced the CD44 expression in mPrE-/- cells. Similar results also occurred when we examined another stem cell marker, Sca1 (Fig. 5). Together, results from Fig. 4-5 suggest that TR4 may be able to increase the prostate cells de-differentiation into the stem cells in the PPARG-deleted cells; then in the in vivo xenografted environment, cells can differentiate into either epithelial or mesenchymal type of tumors.
TR4 increases EMT in PPARG-deleted prostate cells
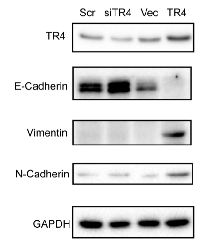
Another way to turn the epithelial cells into the mesenchymal cells is through the epithelial-mesenchymal transition (EMT) [33, 34]. We extracted proteins from the mPrE-/- cell lines to investigate the EMT marker levels. We found E-Cadherin protein increased in the TR4 knocked-down mPrE-/- cells, while it was almost undetectable in the TR4 over-expressed mPrE-/- cells, indicating the strong EMT phenotype (Fig. 6). We further confirmed this EMT via testing other markers and found Vimentin and N-Cadherin were also increased in the TR4 over-expressed mPrE-/- cells (Fig. 6).
Together, results from Figs. 4-6 suggest that TR4 may play key roles to promote the trans-differentiation in the xenografted mice either via altering the stem cell transition and/or EMT in the PPARG-deleted cells.
DISCUSSION
Here we provide the evidences that a nuclear receptor (TR4) impact on PCa progression can be switched based on the availability of another nuclear receptor (PPARG). This is not only unique among nuclear receptors, but it may also have an impact on the future drug development that is based on targeting the nuclear receptors. Early studies suggested that TR4 is a tumor suppressor through regulating the DNA damage/repair system to delay the PCa initiation [27]. Results from current studies, however, demonstrate that TR4 can function as a tumor enhancer via regulating the stem cell population and EMT to promote PCa initiation if PPARG is deleted or its function is suppressed by the GW9662 inhibitor. Both cancer stem cell and/or EMT induced cancer initiation have been reported recently [35-38]. Surprisingly, EMT also contributes to the benign prostatic hyperplasia [39], indicating EMT is not only essential in the later metastatic stage [33, 34], but also plays critical roles in the cell proliferation and cancer initiation.
Why can a single nuclear receptor have these contrasting roles? While the potential explanation could be due to a simple competition between these 2 receptors, the exact mechanism is still elusive and needs further detailed investigations. For instance, under the normal PPARG circumstances, TR4 may function as a tumor suppressor to repair the damaged DNA with little ability to change the stem cell population and/or EMT. However, under the PPARG deleted circumstances (either the gene is lost or loses function), TR4 may still keep its DNA damage repair function but also gain the ability to increase the stem cell population and ability to alter the EMT.
Future clear mechanism dissection of how these 2 receptors interact would resolve the puzzles why TR4 and PPARG have those opposite roles in different diseases including insulin resistance, atherosclerosis, osteoporosis, and now PCa. Importantly, future drugs via targeting either TR4 or PPARG may need extra consideration and be re-examined carefully for additional potential side effects. For example, the anti-diabetic drug TZDs may enhance both PPARG and TR4 activities. However, if the patients have the one allele PPARG deletion, then taking TZD may have the potential risk to promote the PCa development. A better approach will be to examine the PCa patients’ TR4 and PPARG expression in advance before receiving any drugs based on targeting either of these 2 receptors.
MATERIALS AND METHODS
Prostate cancer and paired benign prostate samples
69 cases of human PCa and paired benign appearing prostate specimens were provided by the Surgical Pathology archives after appropriate approval from the Institutional Review Board of the University of Rochester Medical Center.
Fluorescence In situ Hybridization (FISH)
In situ hybridization was performed by the University of Rochester Medical Center Pathology Lab. Green 5-Fluorescein dUTP labeled Human BAC Clone RPCI-11 109P2 probes for human PPARG were purchased from Empire Genomics and we followed their protocol for hybridization.
Cell culture
The spontaneously immortalized mouse prostatic epithelial cell lines, mPrE-/- or mPrE+/+, were generated by Dr. Min Jiang [15] from the PPARG knockout or wild type mouse, respectively. They were maintained in RPMI 1640 media (GIBCO) supplemented with 10% fetal bovine serum and 1% Antibiotic-Antimycotic solution (Invitrogen). The RWPE1 cell line was obtained from the American Type Culture collection (ATCC, Rockwell, MD) and maintained in complete keratinocyte serum-free media (KSF media), supplemented with 1% penicillin/streptomycin/amphoterycin B, 50 mg/ml bovine pituitary extract and 5 ng/ml epidermal growth factor (Life Technologies, Barcelona, Spain). Stable cell lines expressing scramble shRNA (scr) or shRNA (CGGGAGAAACCAAGCAATT) against TR4 (TR4-siRNA) were established by transfecting pcDNA6/TR and pSuperior.retro.puro plasmids into mPrE-/-, mPrE+/+, or RWPE1 cells and selected for stable cell lines by treatment with blasticidin (12 µg/ml) and puromycin (1.2 µg/ml) for two weeks. Tetracycline (1 µg/ml) was added in order to induce shRNA expression. Stable cell lines expressing TR4 or vector control were established by transfecting pcDNA5/TO-TR4 (TR4-cDNA) or pcDNA5/TO (Vec) together with pcDNA6/TR plasmids into mPrE-/- cells and selected for stable cell lines by treatment with blasticidin (12 µg/ml) and hygromycin (100 µg/ml) for two weeks. Tetracycline (1 µg/ml) was added in order to induce TR4 expression.
RNA extraction and quantitative PCR
RNA was extracted from cell lines using TRIzol® reagent (Invitrogen) and converted to cDNA by Superscript III transcriptase (Invitrogen). QPCR was performed using Bio-Rad CFX96 system with SYBR green (Bio-Rad) to determine the level of mRNA expression of a gene of interest. Expression levels were normalized to the expression of β-actin RNA.
Cell proliferation assay
Cells were sub-cultured to 10% confluence then treated with 100 μM N-nitrosomethylurea (NMU) or vehicle control DMSO for 2 hours. Then the cells were grown to reach 100% confluence. Repeat the procedure of NMU treatment for total 3 cycles. After 3 cycles of treatment, passage the cells 5 more times. The treated mPrE-/-, mPrE+/+, or RWPE1 cells were then seeded in 24-well plates (3000 cells/well) and cultured for 24, 48, 72, or 96 hours. After each time point, cell numbers were calculated after using MTT assay staining.
Cell transformation/colony forming assay
Cells were treated with 100 μM NMU or vehicle control DMSO for 3 cycles followed by 5 passages. Cells were suspended at a density of 2×105 cells/ml in 0.4% Noble agar (Sigma, St. Louis, MO) containing media and seeded on top of 0.8% agarose containing media in culture plates plus 2 ml media on top of agar. Media were changed every week for one month. Colonies were stained with p-iodonitrotetrazolium violet (Sigma, St. Louis, MO), photographed, and counted.
Subcutaneous xenograft model
Male 6-8 weeks old nude mice were purchased from NCI. 1x106 cells with matrigel mixture were subcutaneously injected into the nude mice at 8 weeks of age. The tumors were evaluated at 7 or 14 weeks after injection.
Tissue processing and H&E Staining
Xenografted tumor tissues were collected and fixed by 10% formalin followed by paraffin embedding. Samples were sliced to 5 μm thickness followed by hematoxylin and eosin staining.
Cell immunofluorescence staining
105 cells were seeded in 4-well chamber slides. After 24 hours, cells were fixed with acetone for 5 minutes. After 30 minutes blocking, 2 μg/ml of primary antibodies anti-CD44 or anti-Sca1 conjugated with fluorescence were added to the slides overnight at 4°C. Cells were then mounted with mounting solution containing DAPI nucleus staining and pictures were taken with fluorescence microscopy.
Western blot analysis
Target cells were harvested and lysed in RIPA buffer (50 mM Tris–HCl/pH 7.6: 150 mM NaCl: 1% Triton-x-100: 1% sodium deoxycholate: 0.1%SDS) supplemented with complete mini protease inhibitor cocktail tablets. Protein concentration was estimated using the Bio-Rad protein assay (Bio-Rad, Marnes-la-Coquette, France). 50 μg of sample proteins were loaded and run on the 10% SDS-PAGE gel and transferred to PVDF membranes (Millipore). Nonspecific binding was blocked in TBST buffer (0.5 mM Tris-HCl, 45 mM NaCl and 0.05% Tween 20, pH 7.4) containing 5% non-fat powdered milk. Membranes were incubated with primary antibodies anti-E-Cadherin (R&D), anti-Vimentin (Santa Cruz), and anti-N-Cadherin (Abcam) overnight at 4°C. Membranes were then washed in TBST and incubated with HRP conjugated second antibody for 1 hr. The blots were then washed in TBST, followed by incubating with ECL solution according to the manufacturer’s instructions (GE Healthcare).
Statistics
The data values were presented as the mean±SEM. P values were calculated by unpaired Student’s t test or Fisher’s exact test. P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank Karen Wolf for help with manuscript preparation.
Financial support
This work was supported by NIH Grants (CA156700, CA127548 and DK73414); Taiwan Department of Health Clinical Trial and Research Center of Excellence Grant DOH99-TD-B-111-004 (China Medical University, Taichung, Taiwan).
Disclosure Statement
The authors have nothing to disclose.
- 1. Human and rat TR4 orphan receptors specify a subclass of the steroid receptor superfamily. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:6040-4. https://doi.org/10.1073/pnas.91.13.6040. [PubMed].
- 2. Loss of TR4 orphan nuclear receptor reduces phosphoenolpyruvate carboxykinase-mediated gluconeogenesis. Diabetes. 2007;56:2901-9. https://doi.org/10.2337/db07-0359. [PubMed].
- 3. Activation of TR4 orphan nuclear receptor gene promoter by cAMP/PKA and C/EBP signaling. Endocrine. 2009;36:211-7. https://doi.org/10.1007/s12020-009-9220-0. [PubMed].
- 4. Metformin inhibits nuclear receptor TR4-mediated hepatic stearoyl-CoA desaturase 1 gene expression with altered insulin sensitivity. Diabetes. 2011;60:1493-503. https://doi.org/10.2337/db10-0393. [PubMed].
- 5. TR4 is a Tumor Suppressor of Prostate Tumorigenesis via Modulation of DNA Damage/Repair System. Submitted. 2013. https://doi.org/10.1093/carcin/bgu052. [PubMed].
- 6. Subfertility with defective folliculogenesis in female mice lacking testicular orphan nuclear receptor 4. Molecular Endocrinology. 2008;22:858-67. https://doi.org/10.1210/me.2007-0181. [PubMed].
- 7. Stage dependent and androgen inductive expression of orphan receptor TR4 in rat testis. Biochemical and Biophysical Research Communications. 2006;341:464-9. https://doi.org/10.1016/j.bbrc.2005.12.207. [PubMed].
- 8. Abnormal cerebellar cytoarchitecture and impaired inhibitory signaling in adult mice lacking TR4 orphan nuclear receptor. Brain Res. 2007;1168:72-82. https://doi.org/10.1016/j.brainres.2007.06.069. [PubMed].
- 9. The roles of testicular orphan nuclear receptor 4 (TR4) in cerebellar development. Cerebellum. 2008. https://doi.org/10.1007/s12311-008-0006-3. [PubMed].
- 10. Reduced osteoblast activity in the mice lacking TR4 nuclear receptor leads to osteoporosis. Reproductive biology and endocrinology : RB&E. 2012;10:43. https://doi.org/10.1186/1477-7827-10-43. [PubMed].
- 11. TR4 nuclear receptor functions as a fatty acid sensor to modulate CD36 expression and foam cell formation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13353-8. https://doi.org/10.1073/pnas.0905724106. [PubMed].
- 12. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes & Development. 1994;8:1224-34. https://doi.org/10.1101/gad.8.10.1224. [PubMed].
- 13. Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147-56. https://doi.org/10.1016/0092-8674(94)90006-x. [PubMed].
- 14. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585-95. https://doi.org/10.1016/s1097-2765(00)80209-9. [PubMed].
- 15. Disruption of PPARgamma signaling results in mouse prostatic intraepithelial neoplasia involving active autophagy. Cell Death and Differentiation. 2010;17:469-81. https://doi.org/10.1038/cdd.2009.148. [PubMed].
- 16. Yang DR, Lin SJ, Ding XF, Miyamoto H, Messing E, Li LQ, et al. Higher expression of peroxisome proliferator-activated receptor gamma or its activation by agonist thiazolidinedione-rosiglitazone promotes bladder cancer cell migration and invasion. Urology. 2013;81:1109 e1-6. https://doi.org/10.1016/j.urology.2012.12.027. [PubMed].
- 17. PPAR agonists in the treatment of atherosclerosis. Current opinion in Pharmacology. 2003;3:186-91. https://doi.org/10.1016/s1471-4892(03)00014-6. [PubMed].
- 18. Pioglitazone-induced reductions in atherosclerosis occur via smooth muscle cell-specific interaction with PPAR{gamma}. Circulation Research. 2010;107:953-8. https://doi.org/10.1161/CIRCRESAHA.110.219089. [PubMed].
- 19. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496-503. https://doi.org/10.1038/nm1672. [PubMed].
- 20. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J Steroid Biochem Mol Biol. 2002;81:291-308. https://doi.org/10.1016/s0960-0760(02)00118-8. [PubMed].
- 21. Activation of testicular orphan receptor 4 by fatty acids. Biochimica et Biophysica acta. 2009;1789:734-40. https://doi.org/10.1016/j.bbagrm.2009.09.010. [PubMed].
- 22. Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). The Journal of Biological Chemistry. 1995;270:12953-6. https://doi.org/10.1074/jbc.270.22.12953. [PubMed].
- 23. PPARgamma vs TR4 nuclear receptor differential roles in prostate cancer and metabolic diseases. Endocrine-related Cancer. 2014. https://doi.org/10.1530/ERC-13-0529. [PubMed].
- 24. Muscle-specific Pparg deletion causes insulin resistance. Nat Med. 2003;9:1491-7. https://doi.org/10.1038/nm956. [PubMed].
- 25. PPAR gamma and the treatment of insulin resistance. Trends in Endocrinology and Metabolism: TEM. 2000;11:362-8. https://doi.org/10.1016/s1043-2760(00)00306-4. [PubMed].
- 26. Dietary conjugated linoleic acid increases PPAR gamma gene expression in adipose tissue of obese rat, and improves insulin resistance. Growth hormone & IGF research : official journal of the Growth Hormone Research Society and the International IGF Research Society. 2008;18:361-8. https://doi.org/10.1016/j.ghir.2008.01.001. [PubMed].
- 27. TR4 nuclear receptor functions as a tumor suppressor for prostate tumorigenesis via modulation of DNA damage/repair system. Carcinogenesis. 2014. https://doi.org/10.1093/carcin/bgu052. [PubMed].
- 28. Animal models for the study of prostate carcinogenesis. Journal of Cellular Biochemistry Supplement. 1992;16H:89-98. https://doi.org/10.1002/jcb.240501221. [PubMed].
- 29. GW9662, a potent antagonist of PPARgamma, inhibits growth of breast tumour cells and promotes the anticancer effects of the PPARgamma agonist rosiglitazone, independently of PPARgamma activation. Br J Pharmacol. 2004;143:933-7. https://doi.org/10.1038/sj.bjp.0705973. [PubMed].
- 30. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Research. 2010;70:10433-44. https://doi.org/10.1158/0008-5472.CAN-10-2638. [PubMed].
- 31. Kumar SM, Liu S, Lu H, Zhang H, Zhang PJ, Gimotty PA, et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31:4898-911. https://doi.org/10.1038/onc.2011.656. [PubMed].
- 32. SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis. 2013;2:e61. https://doi.org/10.1038/oncsis.2013.23. [PubMed].
- 33. Complex networks orchestrate epithelial-mesenchymal transitions. Nature reviews Molecular Cell Biology. 2006;7:131-42. https://doi.org/10.1038/nrm1835. [PubMed].
- 34. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature Reviews. 2007;7:415-28. https://doi.org/10.1038/nrc2131. [PubMed].
- 35. The role of cancer stem cells in breast cancer initiation and progression: potential cancer stem cell-directed therapies. The Oncologist. 2012;17:1394-401. https://doi.org/10.1634/theoncologist.2012-0163. [PubMed].
- 36. A mathematical model of cancer stem cell driven tumor initiation: implications of niche size and loss of homeostatic regulatory mechanisms. PloS One. 2013;8:e71128. https://doi.org/10.1371/journal.pone.0071128. [PubMed].
- 37. Constitutively activated Stat3 induces tumorigenesis and enhances cell motility of prostate epithelial cells through integrin beta 6. Molecular and Cellular Biology. 2007;27:4444-53. https://doi.org/10.1128/MCB.02404-06. [PubMed].
- 38. Infiltrating macrophages promote prostate tumorigenesis via modulating androgen receptor-mediated CCL4-STAT3 signaling. Cancer Research. 2013;73:5633-46. https://doi.org/10.1158/0008-5472.CAN-12-3228. [PubMed].
- 39. Alonso-Magdalena P, Brossner C, Reiner A, Cheng G, Sugiyama N, Warner M, et al. A role for epithelial-mesenchymal transition in the etiology of benign prostatic hyperplasia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2859-63. https://doi.org/10.1073/pnas.0812666106. [PubMed].
Last Modified: 2016-06-09 20:42:09 EDT

